R package to Estimate parameters for a hawkes model over spatial positions using this c++ implementation.
Requirement
A C++ compiler supporting C++14 is required:
- g++ >= 5.0
- clang++ >= 3.4
- for any other compiler, check that it supports the -std=c++14 flag
R >= 4.3 (the package may works on older version of R)
Installation
You need the package remotes to install hawkesGenomics from this repository
install.packages("remotes")Then you can install hawkesGenomics with this command
remotes::install_git("https://github.com/franckpicard/hawkes_genomics", force = T)Example
We are going to analyse a subset of the replication origin on the chromosme 1 of hg19.
library("hawkesGenomics")preprocess data
beds <- preprocess_bed(
files = c(
system.file("extdata", "oris.bed.gz", package = "hawkesGenomics"),
system.file("extdata", "CGI.bed.gz", package = "hawkesGenomics"),
system.file("extdata", "G4plus.bed.gz", package = "hawkesGenomics")
),
names = c("Oris", "CGI", "G4plus")
)hawkes computation
res <- compute_hawkes_histogram(
files = beds$preprocess_beds,
names = beds$names,
K = 5,
delta = 1e4
)plot data
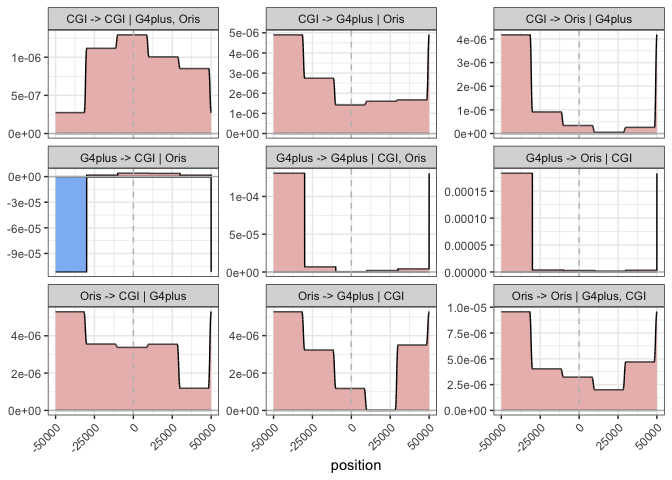
plot_histogram(res, K = 5, delta = 1e4)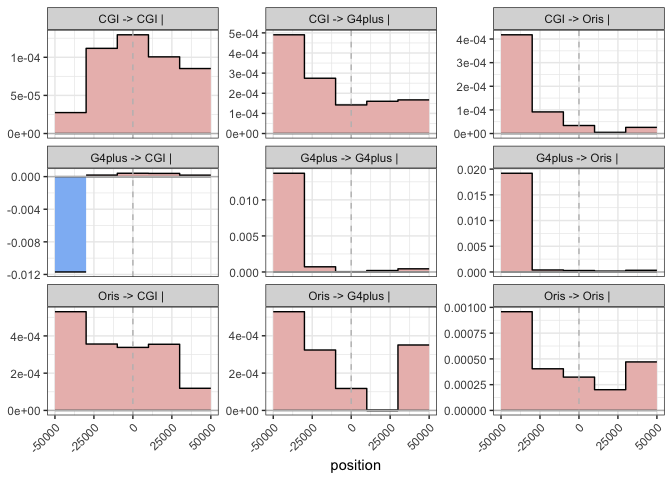
plot_convolution(res, width = beds$interval_size, K = 5, delta = 1e4)